We studied the genetic diversity of indigenous rhizobia nodulating soyabean in South Kivu province of D.R.Congo in order to compare the diversity in grassland and cultivated farms but also to identify indigenous rhizobia with potential of increasing soyabean’BNF and productivity. Soyabean (Glycine max) is an important crop worldwide and especially in Democratic Republic of the Congo, promoted since 1990 to deal with high malnutrition induced by political strife (Kismul et al., 2015).
This study examined the genetic diversity and nitrogen fixation potential of indigenous soyabean nodulating rhizobia (SNR) isolated from cultivated fields and grassland of South Kivu. Seventy SNR isolated from nodules of cultivated and non-cultivated legumes in Uvira, Walungu and Katana territories of South Kivu and a commercial strain (USDA 110) were analysed for genetic diversity based on 16s rRNA, recA, glnII-2 and glnII-12 genes.
Based on all studied genes phylogenies, the results showed that indigenous SNR strains were highly diverse. In addition to the most the reported genus nodulating soyabean Bradyrhizobium and Rhizobium, other strains were identified namely Kosakonia, Bacillus, Beijerinckia, Burkolderia, Microvirga, Cupriviadus, Mesorhizobium and Agrobacterium. The SNR diversity was higher in grasslands compared to cultivated fields. Bradyrhizobium was dominant in cultivated fields (45%) while Kosakonia was dominant in grasslands (35%).
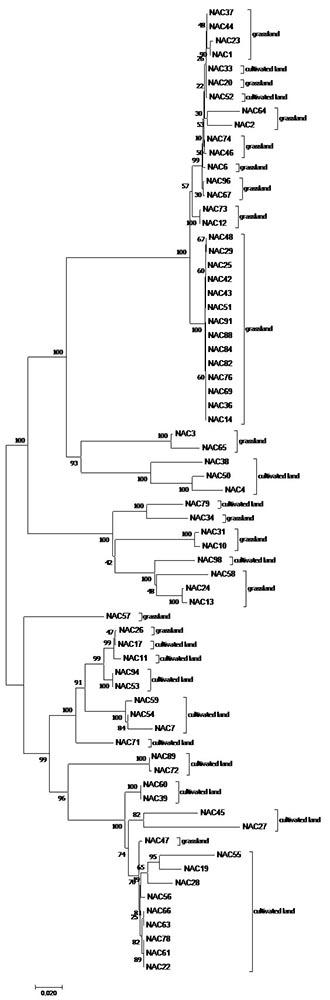
The 16s rRNA phylogeny gave two major clusters divided into five clades whereas the phylogeny based on housekeeping genes (recA and glnII) divided indigenous rhizobia into three well defined clusters in which only Bradyrhizobium (63%) and Rhizobium (37%) were presented. The indigenous SNR isolates (NAC55, NAC76, NAC78, NAC47, NAC61 and NAC35) and USDA 110 clustered together with high bootstrap value (84%) suggesting their high degree of relatedness and possibly high economic values of these indigenous SRN isolates in soyabean production.
|
Figure 1: phylogeny of indigenous SNR strains (NAC) collected from grassland and cultivated fields, based on 16S rRNA gene using the Neighbor-Joining method and the Kimura 2-parameter model modelled by a gamma distribution. The bootstrap values (1000 replicates) are shown next to the branches. |
 |
This study showed that indigenous SNR in South Kivu soils were highly diverse (the computed diversity in the entire population =0.180) and this diversity is even higher in grasslands showing that grassland is an important source of rhizobia. This study showed the presence of promising indigenous rhizobia for improving BNF and soyabean productivity in South Kivu.
This project was supported by the BecA-ILRI Hub through the Africa Biosciences Challenge Fund (ABCF) program. We thank the N2 Africa program for supporting the rhizobia strains collection and maintenance. The Organization for Women in Science for the Developing World (OWSD) is also acknowledged for scholarship grant. The RUFORUM and University of Nairobi are acknowledged for supporting this PhD study.
Ndusha Bintu Nabintu, PhD student University of Nairobi, Kenya. IITA supervisor: Dr. Nabahungu Leon (Click here for her 2018 update)
