| After her MSc within N2Africa Bintu Nabintu Ndusha is enrolled for PhD at University of Nairobi since 2016 and is working under the supervision of Prof. Shellemia Keya and Richard Onwonga of UoN, Dr Leon Nabahungu of IITA and Prof. Gustave Nachigera from her home institute, Université Evangelique en Afrique. She did her Master’s studies under N2 Africa program in 2011 and after her masters she applied for African Bioscience Challenge Fund (ABCF) of BecA-ILRI hub to perform molecular studies on rhizobia isolates obtained by N2Africa in Congo. She obtained CORAF-WECARD/IITA and RUFORUM grants for her field data collection but she is also supported by the Organization of Women in Science for Developing Countries (OWSD). Her PhD project is on genetic diversity and genetic component associated with nitrogen fixation in indigenous rhizobia nodulating soyabean in South Kivu. |

|
Soyabean is an important legume amongst legumes in term of being a crucial source of protein and most consumed edible oil. Apart from its high nutrition value, this crop is capable to fix naturally nitrogen from atmosphere in symbiotic association with bacteria of the Rhizobium genus trough Biological Nitrogen Fixation process. Despite their importance in improving nutrition, food security and soil fertility, soyabean yield in South Kivu remains among the lowest in the world; only 0.5t ha-1. This low yield of soyabean is attributed to a number of factors and the main factors in South Kivu include declining soil fertility accentuated by low use of fertilizers, access to input and improved varieties and poor agronomic practices.
BNF is the best option to improve production of legumes. The use in inoculants of highly effective and adapted rhizobia strains is necessary where the soil does not contain high population of indigenous rhizobia. The gene based-characterization methods are accurate and time saving way to characterize and select high effective rhizobia for inoculants production. The sequencing of genome enables to identify caracteristics associated with compatibility with host and ability of nitrogen fixation in Rhizobium. This method can assess also factors associated with environmental fitness, saprophytic competence and competitiveness for nodule occupancy.
| This study is therefore investigating the genetic diversity and the genetic component associated with N fixation in indigenous rhizobia isolated from South Kivu soil. In addition, the current study will test selected strains for their competitiveness for nodule occupancy and their effectiveness in improving BNF and productivity of soyabean in different soils conditions in South Kivu. For the genetic diversity assessment, three types of genes were used namely: 16s rRNA, housekeeping genes (recA, atpD and glnII) and symbiotic genes (nodA and nifH). Genes used to assess the genetic diversity of indigenous rhizobia are presented in the table 1. To determine the genetic component associated with Nitrogen fixation, the full genome sequencing was performed on Illumina Miseq at BecA-ILRI hub/Kenya. Genomes characteristics were compared to the references genes by the Bidirectional Best Hit (BBH) approach and gene annotation was performed by genes annotation software GLIMMER. The molecular study is complete and the field experiment for indigenous isolates testing for competitiveness is ongoing. | 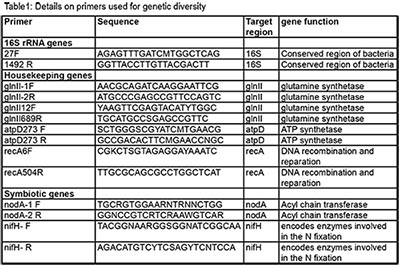 |

|
DNA was extracted from indigenous rhizobia cultures (NAC) obtained from IITA-Kalamo Rhizobiology laboratory. DNA was extracted by QI GEN extraction kit following manufacturer’s instructions. The presence, the absence, the quantity and quality of DNA was assessed using nanodrop method and electrophoresis. The concentration of DNA in samples varied between 20 and 130 ng/µl as indicated in the table 1. The presence of DNA was notified in the majority of samples as indicated on the figure 1. |
Bintu Nabintu Ndusha, University of Nairobi, Kenya (No previous updates)
