Cowpea is an important grain legume valued for its N2-fixing ability and nutritional attributes of its grain and leaves. The symbiotic relationship of cowpea with its microsymbiont rhizobia do account for 96% of its N requirement, can as well contribute to the N needs of subsequent cereal crops in Sub-Saharan Africa, where soils are nutrient-poor. Several studies has indicated that Bradyrhizobium and Rhizobium are the microsymbiont that nodulate cowpea but much remains to be known about the diversity of the naturally occuring rhizobia strains in the major growing cowpea regions in Nigeria. The result explained in this write up show the analysis of the multilocus sequence analysis of 16srRNA in combination with housekeeping gene.
Soil samples were collected from 54 different farm field in three cowpea growing region in Nigeria (Niger, Kaduna, and Kano). Three rhizobia strains were isolated from each soil samples making a total of 162 strains isolated. The strains were subsequently authenticated in further experiment using cowpea cultivar IT97K-499-35 as the trap host to ascertain whether these strains can renodulate cowpea. Subsequently the strains were further amplified via a direct colony polymerase chain reaction using primer pairs for 16S, gyrB, glnII and RecA gene. The purified DNA was sequenced and further phylogenetic analysis were carried out using MEGA 7 software. The multilocus sequence analysis of recA, glnII, gyrB and 16S rRNA were reconstructed using the maximum likelihood method using Kimura’s 2-parameter model. Genospecies were defined based on the MLSA relationship using a 97% sequence similarity threshold.
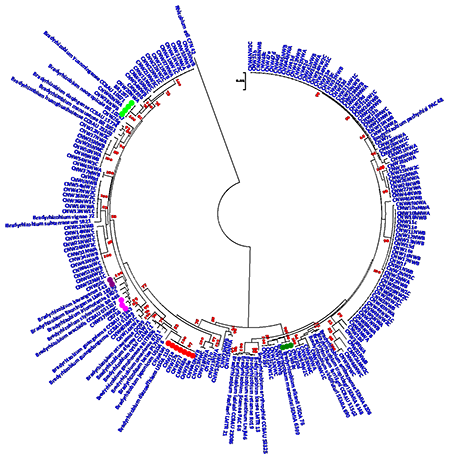
| The isolates shown in Figure 1 clustered with B. elkanii, B. daqingense, B. kavangense, B. yuanmingense and B. diazoefficiens which is indicative of what other studies show. Isolates labelled with red all cluster with B diazoefficiens USDA 110 with 100 % sequence identity. Furthermore, our results also showed that isolate CNW26g clustered with B. daqingense (pink label), a strain originally isolated from soyabean nodules in China and subsequently found to induce effective nodulation on cowpea. The presence of B. daqingense in African soils could be caused by its introduction with soyabean seeds from China. Isolates CNW20d, CNW38c clustered with the B. elkanii group with 100% sequence identity. The 162 rhizobia isolated from cowpea in the three growing regions showed great diversity and biogeographical patterns. |
Figure 1: Maximum likelihood phylogeny of cowpea nodulating rhizobia from Nigeria based on concatenated sequences of 16SrRNA-glnII-gyrB-recA genes |
Ojo Comfort Tinuade, Wageningen University & Research, The Netherlands (Click here for her 2018 update)