The aim of my study is to unravel the types, nature, genetic diversity and effectiveness of groundnut-nodulating rhizobia existing in different soil types of the moist; northern Guinea (NG) and dry; Sudan (S) savannas of Nigeria, with a view to identify strains that hold promise for improved symbiotic N2-fixation, productivity and soil fertility. After the last report of my studies in 2015 (Podcaster 32), there have been significant progress and fascinating findings with regard to the genetic diversity of groundnut-nodulating rhizobia. The 32 novel groundnut-nodulating rhizobia strains from the Nigerian NG and S savannas, isolated in the course of my PhD study, were subjected to RPO1 finger printing (Richardson et al., 1995). Overall, the results showed very high genetic diversity among the strains.
Eleven representatives of the novel Nigerian indigenous groundnut-nodulating rhizobia strains were selected, based on growth of the strains on YMA media and subjected to 16S rDNA sequencing, which further confirmed the strains’ genetic diversity, earlier shown by the RPO1 finger printing. Over 90% of them are Bradyrhizobium strains, all closely matching (query cover 98-100% and identity 99-100%) with varying species deposited in the genebank. The strains were tested for effectiveness in the glasshouse on an Australian groundnut cultivar (Kangaroy peanut). The results indicated compatibility and high effectiveness of the strains on the cultivar, indicating their potential for adoption as commercial inoculant strains within Nigeria and beyond.
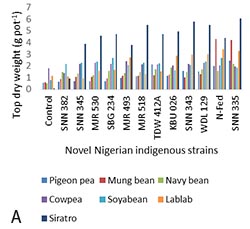
Furthermore, the strains were subjected to host range studies on seven legume hosts; navy bean (Phaseolus vulgaris L.), mungbean (Vigna radiata), soyabean (Glycine max), siratro (Macroptilium atropurpureum), pigeon pea (Cajanus cajan Millsp), cowpea (Vigna unguiculata) and lablab (Lablab purpureus). These host range studies were conducted to test the level of promiscuity among the strains and the legume hosts. The results showed high level of promiscuity among both the strains and the legume hosts. Most of the strains effectively nodulated with all the legume hosts. The exception was navy bean, which was nodulated by all the strains, but none was effective. SNN 382 did not nodulate siratro, mungbean and pigeon pea. This strain also had the least influence on top dry weight of the legume hosts (Figure 1A). Significant interaction was observed between the strains and the hosts in top dry weight (g pot-1), relative to the uninoculated control (Figure 1A).
 |
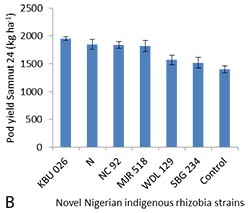 |
Figure 1. (A) Interaction between the eleven representatives of the Nigerian indigenous rhizobia strains and seven host legumes in top dry weight (g pot-1) relative to uninoculated control and (B) combined performance of selected novel Nigerian indigenous rhizobia strains in both northern Guinea and Sudan savannas of Nigeria, as tested in the research farms of the Institute for Agricultural Research, Ahmadu Bello University, Zaria Nigeria at Samaru and Minjibir during the 2015/2016 growing season |
Field experiments in the Nigerian NG and S savannas, the original environments from which the strains were isolated, using four of the strains which have been confirmed effective in earlier reported glasshouse and field experiments (Abdullahi et al., unpublished but presented in Podcaster 32), indicated KBU 026 to be the best among the strains, with mean pod yield of 1,953 kg ha-1 obtained from combined analysis of data for both the NG and S savannas. The strain performed significantly better than the N-fed control (20 N kg ha-1), all other strains (including NC 92) and the uninoculated control (other competing native rhizobia strains in the field) (Figure 1B). This shows the high potential of the strains, particularly KBU 026 for adoption as an inoculant strain. I am currently in Nigeria and at high stage of my thesis writing, in contact with my supervisors, hoping to complete my studies as soon as possible.
Aliyu Anchau Abdullahi, Murdoch University, Australia
Reference: Richardson, A. E., Viccars, L. A., Watson, J. M. and Gibson, A. H. (1995). Differentiation of Rhizobium strains using the polymerase chain reaction with random and directed primers. Soil Biology and Biochemistry, 27 (4/5) pp. 515-524.
