Despite decades of intensive research on Rhizobium-legume symbioses, we are still only beginning to understand the factors that influence the survival, persistence, efficiency and competition of rhizobial strains in various environmental conditions.
Research focused on the identification of both competitive and highly effective nitrogen-fixing strains has been limited because, so far, the only way to test competitive rhizobial strains has been to isolate individual strains and compare them one at a time in large-scale plant growth assays.
Four years ago, I began my PhD in Professor Philip Poole’s lab (https://rhizosphere.org/) with the aim of engineering a molecular toolkit for high-throughput identification of competitive and effective rhizobial strains.
|
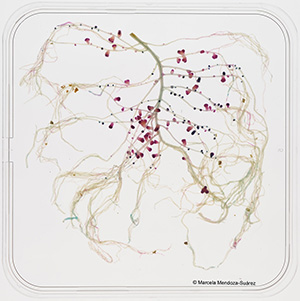
We have developed a library of novel reporter plasmids (Plasmid IDs) based on the broad host range vector pOGG026 that is suitable for rhizobial screening in the rhizosphere (Geddes et al., 2019). The Plasmid IDs contain the maker gene sfGFP under the control of a synthetic consensus promoter (PsnifH), which drives nodule-specific expression in biovars and strains of Rhizobium leguminosarum and which can be used to assess competitiveness of strains without reducing their fitness and as a proxy for nitrogenase activity assays. The Plasmid IDs also contain specific nucleotide sequences so that, in a single experiment, multiple rhizobial strains can be marked and scored for nodule occupancy through the use of next generation sequencing (NGS). Additionally, we developed reporter plasmids that express the biomarkers gusA and celB, allowing us to develop a new plasmid-based system to mark Rhizobium strains and evaluate their competitiveness (Fig. 1). |
Figure 1. Competition assays for nodule formation between UPM791-gusA under the control of PsnifH (pink nodules) and Rlv3841- celB under the control of PsnifH (blue nodules) in pea roots. |
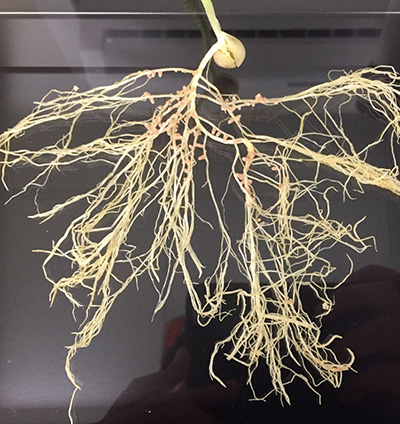
The application of this tool in a competition assay among eighty-four Tagged-strains, in non-sterilised agricultural soil containing indigenous rhizobia (Fig. 2), allowed us to identify an effective and super competitive strain. These results demonstrated the proof of concept of Plasmid IDs as a tool for high-throughput identification of competitive and effective rhizobial strains.
 |
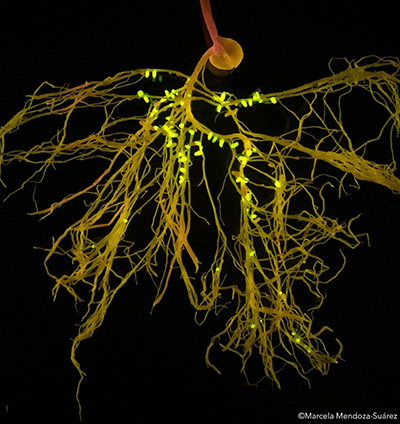 |
Figure 2. (Left) Photograph obtained using a mobile phone camera; (Right) Roots were exposed to a Safe blue-light transilluminator and the mobile phone camera was accompanied by an orange filter. Nodules expressing green fluorescence occupied by Tagged-strains with Plasmid-IDs and non-fluorescent nodules occupied by indigenous rhizobia.
This tool has the potential to revolutionise the search for elite indigenous rhizobia, by reducing the costs involved in identifying competitive and effective rhizobial strains. It provides a fantastic opportunity to identify indigenous rhizobial strains with both competitive and effective traits, and then use them in similar soil types where ineffective rhizobial strains are found.
Marcela Mendoza-Suárez, Post-doctoral Researcher, Department of Plant Sciences, University of Oxford, UK
Reference:
Geddes, B. A., Mendoza-Suárez, M. A., and Poole, P. S. (2019). A Bacterial Expression Vector Archive (BEVA) for Flexible Modular Assembly of Golden Gate-Compatible Vectors. Front. Microbiol. 9, 3345. doi:10.3389/fmicb.2018.03345.
Funding:
This work was supported by the Biotechnology and Biological Sciences Research Council (Grant No. BB/L011484/1), the Mexican government (CONACYT-I2T2 Nuevo León Grant code 266954/399852) and Oxford Balliol College (Foley-Béjar scholarship).