Introduction: Chickpea (Cicer arietinum L.) is one of the grain legumes that symbiotically interact with Mesorhizobium strains and fix atmospheric N. The efficiency of the symbiotic N fixation partly depends on the host genotype (GL) and rhizobium strains (GR). Here, we studied GL x GR interaction between genetically determined Mesorhizobium strains and chickpea genotypes primarily in jars and subsequently in pots in Ethiopia.
Methodology: chickpea genotypes (11GL) were selected from previously described accessions (Updhayaya, unpublished) based on genetic distances between groups1 and imported from ICRISAT-India. Five reference strains (reported to nodulate chickpea and vary genetically2,3, imported from LMG strain collection centre, Belgium) and five phylogenetically characterized local strains following protocols described earlier (see my 2018 update) were selected and studied. Factorial combinations of 11GL x 12 (10GR + 2 controls (0.05g/ml N fertilized and unfertilized) with 5 replications were tested in jars on sterile sand with RCBD. All treatments were supplemented with Jenson’s N-free nutrient solution and assessed for symbiotic effectiveness after 45 days of growth in screening house. Subsequently, 5GL x 8 (6GR + 2 controls) combinations were tested in pots with similar design as in jars. Nodulation and fixation were measured as nod+/- and fix+/- respectively and visualized to detect patterns (if any) using heatmaps in R version 3.5. Furthermore, symbiotic effectiveness was analysed based on shoot biomass and fixed N using mixed linear models. Combinations with higher symbiotic performance were assessed using additive main effect and multiplicative interaction (AMMI) model in R using agricolae package.
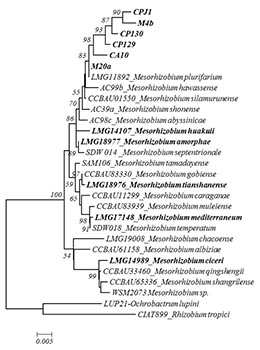
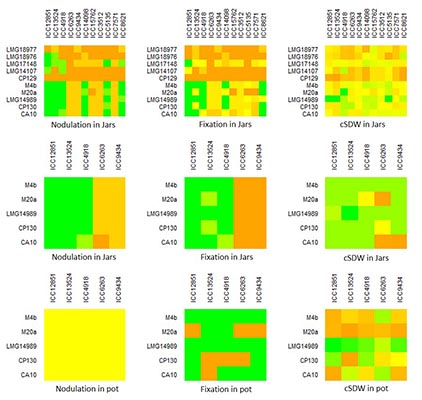
Results: Phylogenetic analysis of the reference and local strains, based on 16s rRNA sequences, showed genetic breadth (Fig. 1). However, all the local strains were assigned to M. plurifarium (a microsymbiont of Acacia senegal L. but sporadically found in chickpea root nodules4,5) at lower bootstrap (83%) while the reference strains were evenly distributed in the entire tree. Nodulation and fixation genes grouped the local strains with M. ciceri and M. mediterraneum (data not shown), reflecting different evolutionary roots in symbiotic and 16s rRNA genes. Heatmaps revealed some patterns in terms of nodulation, fixation and plant shoot biomass (Fig. 2). The distinct and consistent patches indicated widely in the jar and jar subset (the upper three and middle three heatmaps in Fig. 2) were not the real differences observed among the combinations but were due to the missing values in the jar recorded for Kabuli genotypes as a result of weak germination. Besides this problem, weak patterns are observed when only desi genotypes are considered and that is mostly brought about by one strain (M. ciceri, LMG14989). This strain was clearly confirmed to be the most effective one across genotypes in the subsequent study in pots (Fig. 2). It fixed the highest amount of N and induced the highest shoot biomass with all the genotypes among the studied strains and was found to be stable across the genotypes.
|
Fig. 1. Neighbour joining 16s rRNA phylogeny of the reference and local strains. Strains labelled in bold are included in this study. |
Fig 2. Nodulation, fixation and shoot biomass patterns of GL x GR in chickpea. Where, cSDW is corrected shoot dry weight; Chickpea genotypes are indicated on the x-axis while strains are indicated on the y-axis. |
The local strains had weak and similar performance across genotypes, and this is concordant with their similarities reflected in the phylogenetic tree (Fig 1), indicating that they are identical and sporadic symbionts of chickpea that might have obtained symbiotic genes from M. ciceri or M. mediterraneum strains. Practically, this means that the weakly performing local strains (trapped from different locations) dominate in Ethiopian soils and possibly minimize the potential N2-fixation by chickpea, as they are not the distinguished chickpea microsymbionts. Besides, we found evidence for the presence of GL x GR interaction in chickpea in jars (p<0.001), due to effects of Kabuli genotypes. This evidence was disappeared in the pot experiment (data not shown), in which case excess Kabuli seeds were used to replace the missing ones. Thus, pots can be taken as probable confirmatory supporting media for investigating the presence of GL x GR interaction in legumes. An interesting observation is that strain LMG14989 is a broad genotype spectrum strain that fixes significantly higher N and enhances plant growth.
Ashenafi Hailu Gunnabo, Wageningen University & Research, The Netherlands
References:
- van Heerwaarden, J. et al. Genetic signals of origin, spread, and introgression in a large sample of maize landraces. PNAS 108, 1088–1092 (2011).
- Rivas, R. et al. Strains of Mesorhizobium amorphae and Mesorhizobium tianshanense, carrying symbiotic genes of common chickpea endosymbiotic species, constitute a novel biovar (ciceri) capable of nodulating Cicer arietinum. Lett. Appl. Microbiol. 44, 412–418 (2007).
- Laranjo, M., Young, J. P. W. & Oliveira, S. Multilocus sequence analysis reveals multiple symbiovars within Mesorhizobium species. Syst. Appl. Microbiol. 35, 359–367 (2012).
- Laranjo, M. et al. Chickpea rhizobia symbiosis genes are highly conserved across multiple Mesorhizobium species. FEMS Microbiol. Ecol. 66, 391–400 (2008).
- Rivas, R., Garcia-Fraile, P. & Velazquez, E. Taxonomy of bacteria nodulating legumes. Microbiol. Insights 2, 51–69 (2009).