Legumes are capable of establishing symbiotic associations with rhizobia in a process called biological nitrogen fixation which is responsible for the wide adoption of legumes as food crops, forages, green manures and in forestry. Recently, the call for the return to a sustainable form of agriculture due to pollution of water body by nitrate and increase in cost fertilizer has caused an increased interest in biological N2-fixation. Therefore, usage of N2-fixing legumes is not limited to only cash crop or biofuel but could be important in the recovery and improvement of soil fertility. Therefore it is timely to obtain new information regarding legumes and diazotrophic symbiotic bacteria. The process leading to the identification of effective rhizobia strains for cowpea could be labour and time consuming and often result into production of several rhizobia cultures. Therefore, it is imperative we charactize them genetically. This study was carried out to test the discriminatory power and reproducibility of BOX-PCR for fingerprinting rhizobia.
Rhizobia strains were diluted in sterile water and amplified by PCR with primer BOX A1R (5′-CTACGGCAAGGCG ACGCTGACG-3′). These strains included isolated indigenous strains. The amplified fragments were separated by horizontal electrophoresis on 2% agarose gel, stained with ethidium bromide, visualized under UV radiation and photographed. The sizes of the fragments were normalized according to the sizes of the DNA markers. Cluster analyses of the BOX-PCR profiles were performed using GelJ program (version 1.0) with the UPGMA algorithm and the Jaccard coefficient, considering the optimum values indicated by the GelJ program for the tolerance and optimization parameters.
Well defined groups with a high level of inter and intra specific diversity and complex fingerprinting patterns with multiple distinct bands of various intensities were obtained in the BOX-PCR analysis of the rhizobia strains from the different agroecological zones (Figure 1). Images for Niger and Kano Agro ecological zones not shown). None of the strains showed identical (100% similarity) fingerprints, but 99% of similarity or higher were obtained for strains CNW26NWB & CNW25NWA (99.3%), CNW33f & CNW33e (99%)-Kaduna and CNW49NW1C & CNW49NW2C (99.1%)-Kano. In the cluster analysis of the BOX-PCR profiles, strains from each agroecological zone were grouped at similarity levels; Niger 60% (15 groups), Kaduna 58% (37 groups) shown in figure 1, Kano 57% (43 groups).
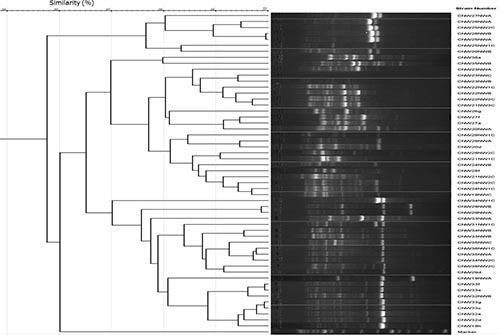
|
Consensus sequences such as BOX related to repetitive and conservative elements diffused in DNA have been extensively used in ecology, genetic and taxonomic studies as well as for rhizobia strain identification and the technique has also proven to be valuable in studies of rhizobia isolated from tropical soils. As shown from the result, other than detection of genetic diversity, the BOX fingerprinting tool was able to show the percentage level of similarity among the different strains from each agro ecological zones. This obtained information might be important when considering the same set of rhizobia strains for phylogenetic analysis of the 16SrRNA region in their genome. Also there will be need to test the effectiveness of these strains in the greenhouse so as to decipher the pattern of N2-fixation with their host cowpea.
Comfort Ojo Tinuade, Wageningen University & Research, The Netherlands (Click here for her 2016 update)
